Prof. Dr. Tobias Madl
Bringing order to protein disorder

Research
“Function requires structure” – this is a common statement throughout structural biology. Indeed, most proteins need to adopt a defined three-dimensional structure to carry out their function. However, a large fraction of the genome of any organism encodes proteins that, completely or in part, do not adopt a defined three-dimensional structure but are nevertheless essential for cellular function: the so-called intrinsically or natively disordered proteins/regions (IDP/Rs). Given their frequent occurrence in disease it is of no doubt that these proteins are preferred targets in drug development. To understand these proteins in general and to provide strategies how to modulate these interactions for disease treatment, detection of structural properties of IDP/Rs and structural characterization of their biological complexes is the key.
My BioSysNet-funded research centers on the key IDP/Rs in the intersection of the Wnt and insulin signaling pathways which regulate cell fate, and contribute to diseases and ageing. Recent work has shown that regulation of these proteins involves an intricate ‘code’ of numerous post-translational modifications (PTMs) and binding of a plethora of co-factors (i.e. ß-catenin) within their IDRs. Deregulation of these proteins, including missense mutations and changes in the signaling state, leads to cancer, diabetes and ageing. Despite their importance, the underlying molecular mechanisms are still largely unknown. This is mainly due to their intrinsic flexibility, which complicates structural analysis by conventional approaches.
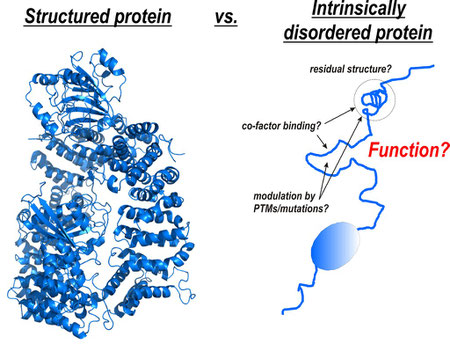
To address this problem I apply my recently developed multidisciplinary approach to characterize the structural properties of these disease-related IDRs in isolation and in complex with their folded protein co-factors, the modulation of residual structure by modifications (PTMs, mutation), and the interference with small-molecule modulators with function in vitro and in vivo. This holds the key to understanding these proteins in general and provides essential inside how to modulate these interactions for disease treatment.
Group members
Dr. Christoph Göbl
Christoph Hartlmüller
Dr. Henry Hocking
Dr. Benjamin Bourgeois
Dr. Sandra Scanu
Publications within BioSysNet
Huang JR, Warner LR, Sanchez C, Gabel F, Madl T, Mackereth CD, Sattler M, Blackledge M (2014). Transient electrostatic interactions dominate the conformational equilibrium sampled by multidomain splicing factor U2AF65: a combined NMR and SAXS study. J Am Chem Soc 136(19):7068-76.
Lorenz OR, Freiburger L, Rutz DA, Krause M, Zierer BK, Alvira S, Cuéllar J, Valpuesta JM, Madl T, Sattler M, Buchner J (2014). Modulation of the Hsp90 chaperone cycle by a stringent client protein. Mol Cell 53(6):941-53.
Göbl C, Madl T, Simon B, Sattler M (2014). NMR approaches for structural analysis of multidomain proteins and complexes in solution. Prog Nucl Magn Reson Spectrosc 80:26-63.
Müller R, Gräwert MA, Kern T, Madl T, Peschek J, Sattler M, Groll M, Buchner J (2013). High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation. High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation. Proc Natl Acad Sci U S A 110(25):10183-8.
Putker M, Madl T, Vos HR, de Ruiter H, Visscher M, van den Berg MC, Kaplan M, Korswagen HC, Boelens R, Vermeulen M, Burgering BM, Dansen TB (2013). Redox-dependent control of FOXO/DAF-16 by transportin-1. Mol Cell 49(4):730-42.
Dormann D, Madl T, Valori CF, Bentmann E, Tahirovic S, Abou-Ajram C, Kremmer E, Ansorge O, Mackenzie IR, Neumann M, Haass C (2012). Arginine methylation next to the PY-NLS modulates Transportin binding and nuclear import of FUS. EMBO J 31(22):4258-75.
Hocking HG, Zangger K, Madl T (2013). Studying the structure and dynamics of biomolecules by using soluble paramagnetic probes. Chemphyschem 14(13):3082-94.
Karagöz GE, Duarte AM, Akoury E, Ippel H, Biernat J, Morán Luengo T, Radli M, Didenko T, Nordhues BA, Veprintsev DB, Dickey CA, Mandelkow E, Zweckstetter M, Boelens R, Madl T, Rüdiger SG (2014). Hsp90-Tau Complex Reveals Molecular Basis for Specificity in Chaperone Action. Cell 156 (5):963-974.
Zhang Y, Madl T, Bagdiul I, Kern T, Kang HS, Zou P, Mäusbacher N, Sieber SA, Krämer A, Sattler M (2013). Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition. Nucleic Acids Res 41(2):1343-54.
Publications before BioSysNet
1. Tavanez J.P., Madl T., Kooshapur H., Sattler M., Valcárcel J., hnRNP A1 proofreads 3’ splice site recognition by U2AF, Molecular Cell (2012) , accepted
2. Tripsianes K., Madl T., Machyna M., Fessas D., Englbrecht C., Fischer U., Neugebauer K.M., Sattler M., Structural basis for dimethyl-arginine recognition by the Tudor domains of SMN and SPF30, Nature Structural and Molecular Biology 18 (2011), 1414-1420
3. Mackereth C.D., Madl T., Bonnal S., Simon B., Zanier K., Gasch A., Rybin V., Valcárcel J. and Sattler M., Multi-domain conformational selection underlies polypyrimidine recognition by U2AF and pre-mRNA splicing, Nature 475 (2011), 408-411
4. Madl T., Güttler T., Görlich D., Sattler M., Structural Analysis of Large Protein Complexes using Solvent Paramagnetic Relaxation Enhancements, Angewandte Chemie 50 (2011), 3993-3997
5. Madl T., Gabel F., Sattler M., NMR and Small Angle Scattering-based structural analysis of protein complexes in solution, Journal of Structural Biology 173 (2011), 472-482
6. Güttler T., Madl T. (shared first author), Neumann P., Deichsel D., Corsini L., Monecke T., Ficner R., Sattler M., Görlich D., Structural basis for the recognition of diverse nuclear export signals by the exportin CRM1, Nature Structural and Molecular Biology 17 (2010), 1367-1376
7. Meyer H., Tripsianes K., Vincendeau M., Madl T., Kateb F., Brack-Werner R., Sattler M., Structural basis for homodimerization of the Src-associated during mitosis, 68 kD protein (Sam68) Qua1 domain, Journal of Biological Chemistry 285 (2010), 28893-28901
8. Madl T., Felli I.C., Bertini I., Sattler M., Structural analysis of protein interfaces from 13C direct-detected paramagnetic relaxation enhancements, Journal of the American Chemical Society 132 (2010), 7285-7287
9. Simon B., Madl T., Mackereth C.D., Nilges M., Sattler M., An Efficient Protocol for NMR-Spectroscopy-Based Structure Determination of Protein Complexes in Solution, Angewandte Chemie 49 (2010), 1967-1970
10. Spirk S., Belaj F., Madl T., Pietschnig R., A Radical Approach to Hydroxylaminotrichlorosilanes: Synthesis, Reactivity, and Crystal Structure of TEMPO-SiCl3 (TEMPO = 2,2,6,6-Tetramethylpiperidine-N-oxyl), European Journal of Inorganic Chemistry 2 (2010), 289-297
11. Madl T., Bermel W., Zangger K.: Use of Relaxation Enhancements in a Paramagnetic Environment for the Structure Determination of Proteins Using NMR Spectroscopy, Angewandte Chemie 48 (2009), 8259-8262
12. Madl T., Sattler M.: Adhesion dance with raver, Structure 17 (2009), 781-783
13. Zangger K., Respondek M., Göbl C., Hohlweg W., Rasmussen K., Grampp G., Madl T.: Positioning of micelle-bound peptides by paramagnetic relaxation enhancements, Journal of Physical Chemistry B 113 (2009) 4400-4406
14. Traar P., Rumpler A., Madl T., Saischek G., Francesconi KA.: Synthesis of Naturally Occurring Arsenic-Containing Carbohydrates, Australian Journal of Chemistry 6 (2009), 538-545
15. Neufeld C., Filipp FV., Simon B., Neuhaus A., Schüller N., David C., Kooshapur H., Madl T., Erdmann R., Schliebs W., Wilmanns M., Sattler M..: Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19, EMBO Journal 28 (2009) 745-754
16. Spirk S., Madl T., Pietschnigg R., Relax with TEMPO: A Paramagnetic Relaxation Agent Useful also for Silicon-29 NMR Spectroscopy, Organometallics 27 (2009), 500-502
17. Respondek M., Madl T., Gobl C., Golser R., Zangger K., Mapping the Orientation of Helices in Micelle-Bound Peptides by Paramagnetic Relaxation Waves, Journal of the American Chemical Society 129 (2007), 5228-5234
18. Madl T., Van Melderen L., Mine N., Respondek M., Oberer M., Keller W., Khatai L., Zangger K., Structural basis for nucleic acid and toxin recognition of the bacterial antitoxin CcdA, Journal of Molecular Biology 364 (2006), 170-185
19. Madl T., Sterk H., Rechberger G.N., Mittelbach M., Tandem mass spectrometric analysis of a complex triterpene saponin mixture of Chenopodium quinoa, Journal of the American Society for Mass Spectrometry 17 (2006), 795-806
20. Madl T., Mittelbach M., Quantification of primary fatty acid amides in commercial tallow and tallow fatty acid methyl esters by HPLC-APCI-MS, The Analyst 130 (2005), 565-570
Journal articles (not peer-reviewed):
1. Madl T., Sattler M., Strukturaufklärung von Proteinkomplexen in Lösung, Laborpraxis (2010)
Book chapters:
1. Madl T., Sattler M., NMR tools and the analysis of protein-protein interactions, in: NMR in Mechanistic Systems Biology, in press



